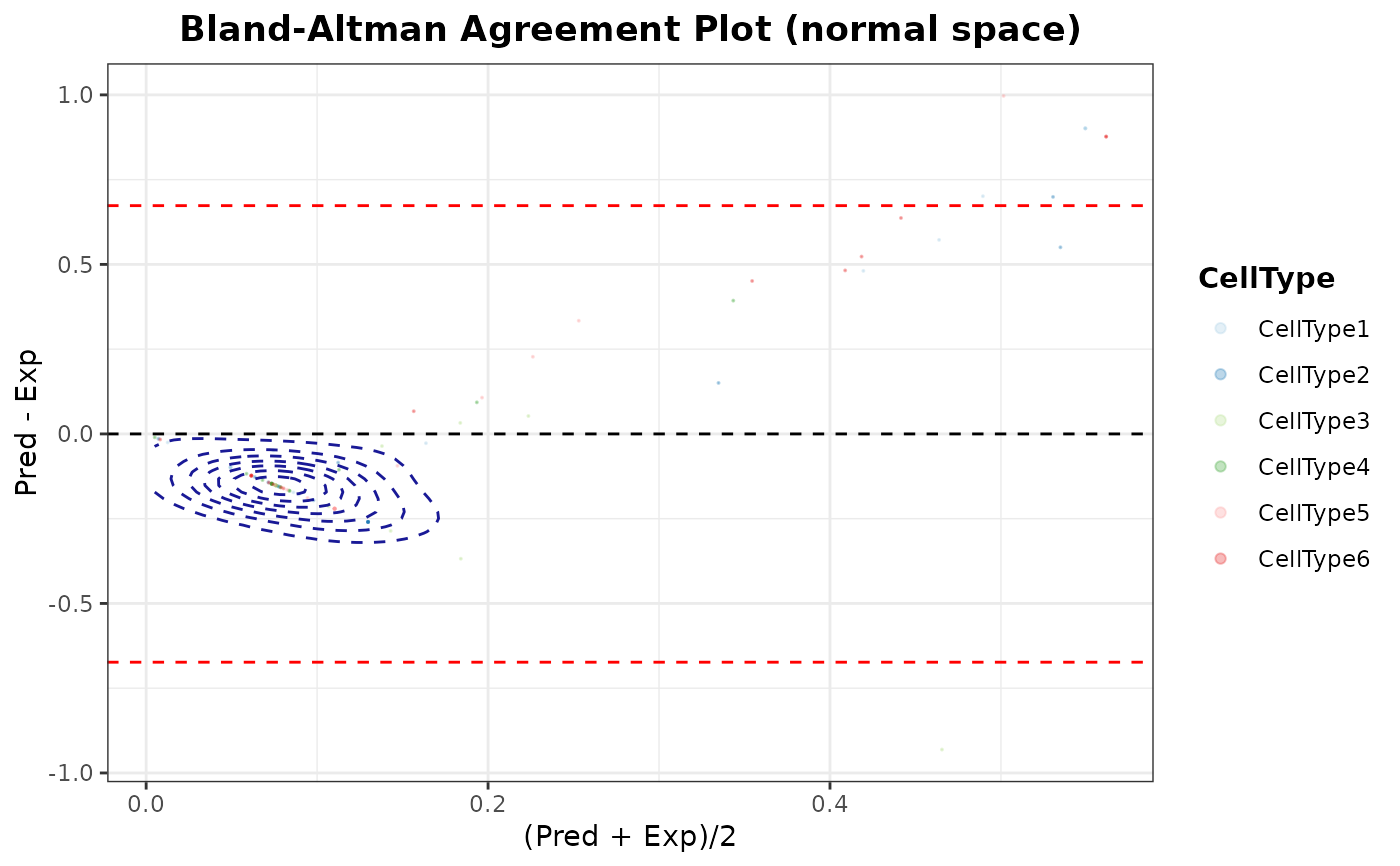
Generate Bland-Altman agreement plots between predicted and expected cell type proportions of test data
Source:R/evalMetrics.R
blandAltmanLehPlot.RdGenerate Bland-Altman agreement plots between predicted and expected cell
type proportions from test data. The Bland-Altman agreement plots can be
shown all mixed or split by either cell type (CellType) or the number
of cell types present in spots (nCellTypes). See the facet.by
argument and examples for more information.
Usage
blandAltmanLehPlot(
object,
colors,
color.by = "CellType",
facet.by = NULL,
log.2 = FALSE,
filter.sc = TRUE,
density = TRUE,
color.density = "darkblue",
size.point = 0.05,
alpha.point = 1,
ncol = NULL,
nrow = NULL,
title = NULL,
theme = NULL,
...
)Arguments
- object
SpatialDDLSobject withtrained.modelslot containing metrics in thetest.deconv.metricsslot of aDeconvDLModelobject.- colors
Vector of colors to be used.
- color.by
Variable used to color data. Options are
nCellTypesandCellType.- facet.by
Variable used to show the data in different panels. If
NULL, the plot is not split into different panels. Options arenCellTypes(by number of different cell types) andCellType(by cell type).- log.2
Whether to show the Bland-Altman agreement plot in log2 space (
FALSEby default).- filter.sc
Boolean indicating whether single-cell profiles are filtered out and only correlations of results associated with mixed spot profiles are shown (
TRUEby default).- density
Boolean indicating whether density lines should be shown (
TRUEby default).- color.density
Color of density lines if the
densityargument isTRUE.- size.point
Size of the points (0.1 by default).
- alpha.point
Alpha of the points (0.1 by default).
- ncol
Number of columns if
facet.byis used.- nrow
Number of rows if
facet.byis used.- title
Title of the plot.
- theme
ggplot2 theme.
- ...
Additional argument for the
facet_wrapfunction of ggplot2 iffacet.byis notNULL.
Examples
# \donttest{
set.seed(123)
sce <- SingleCellExperiment::SingleCellExperiment(
assays = list(
counts = matrix(
rpois(30, lambda = 5), nrow = 15, ncol = 20,
dimnames = list(paste0("Gene", seq(15)), paste0("RHC", seq(20)))
)
),
colData = data.frame(
Cell_ID = paste0("RHC", seq(20)),
Cell_Type = sample(x = paste0("CellType", seq(6)), size = 20,
replace = TRUE)
),
rowData = data.frame(
Gene_ID = paste0("Gene", seq(15))
)
)
SDDLS <- createSpatialDDLSobject(
sc.data = sce,
sc.cell.ID.column = "Cell_ID",
sc.gene.ID.column = "Gene_ID",
sc.filt.genes.cluster = FALSE
)
#> === Spatial transcriptomics data not provided
#> === Processing single-cell data
#> - Filtering features:
#> - Selected features: 15
#> - Discarded features: 0
#>
#> === No mitochondrial genes were found by using ^mt- as regrex
#>
#> === Final number of dimensions for further analyses: 15
SDDLS <- genMixedCellProp(
object = SDDLS,
cell.ID.column = "Cell_ID",
cell.type.column = "Cell_Type",
num.sim.spots = 50,
train.freq.cells = 2/3,
train.freq.spots = 2/3,
verbose = TRUE
)
#>
#> === The number of mixed profiles that will be generated is equal to 50
#>
#> === Training set cells by type:
#> - CellType1: 3
#> - CellType2: 1
#> - CellType3: 3
#> - CellType4: 2
#> - CellType5: 3
#> - CellType6: 2
#> === Test set cells by type:
#> - CellType1: 1
#> - CellType2: 1
#> - CellType3: 1
#> - CellType4: 1
#> - CellType5: 1
#> - CellType6: 1
#> === Probability matrix for training data:
#> - Mixed spots: 34
#> - Cell types: 6
#> === Probability matrix for test data:
#> - Mixed spots: 16
#> - Cell types: 6
#> DONE
SDDLS <- simMixedProfiles(SDDLS)
#> === Setting parallel environment to 1 thread(s)
#>
#> === Generating train mixed profiles:
#>
#> === Generating test mixed profiles:
#>
#> DONE
# training of DDLS model
SDDLS <- trainDeconvModel(
object = SDDLS,
batch.size = 15,
num.epochs = 5
)
#> === Training and test from stored data
#> Using only simulated mixed samples
#> Using only simulated mixed samples
#> Model: "SpatialDDLS"
#> _____________________________________________________________________
#> Layer (type) Output Shape Param #
#> =====================================================================
#> Dense1 (Dense) (None, 200) 3200
#> _____________________________________________________________________
#> BatchNormalization1 (BatchNorm (None, 200) 800
#> _____________________________________________________________________
#> Activation1 (Activation) (None, 200) 0
#> _____________________________________________________________________
#> Dropout1 (Dropout) (None, 200) 0
#> _____________________________________________________________________
#> Dense2 (Dense) (None, 200) 40200
#> _____________________________________________________________________
#> BatchNormalization2 (BatchNorm (None, 200) 800
#> _____________________________________________________________________
#> Activation2 (Activation) (None, 200) 0
#> _____________________________________________________________________
#> Dropout2 (Dropout) (None, 200) 0
#> _____________________________________________________________________
#> Dense3 (Dense) (None, 6) 1206
#> _____________________________________________________________________
#> BatchNormalization3 (BatchNorm (None, 6) 24
#> _____________________________________________________________________
#> ActivationSoftmax (Activation) (None, 6) 0
#> =====================================================================
#> Total params: 46,230
#> Trainable params: 45,418
#> Non-trainable params: 812
#> _____________________________________________________________________
#>
#> === Training DNN with 34 samples:
#>
#> === Evaluating DNN in test data (16 samples)
#> - loss: 1.807
#> - accuracy: 0.0625
#> - mean_absolute_error: 0.2502
#> - categorical_accuracy: 0.0625
#>
#> === Generating prediction results using test data
#> DONE
# evaluation using test data
SDDLS <- calculateEvalMetrics(object = SDDLS)
# Bland-Altman plot by cell type
blandAltmanLehPlot(
object = SDDLS,
facet.by = "CellType",
color.by = "CellType"
)
#> Warning: `stat_contour()`: Zero contours were generated
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: `stat_contour()`: Zero contours were generated
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: `stat_contour()`: Zero contours were generated
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> Warning: `stat_contour()`: Zero contours were generated
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
 # Bland-Altman plot of all samples mixed
blandAltmanLehPlot(
object = SDDLS,
facet.by = NULL,
color.by = "CellType",
alpha.point = 0.3,
log2 = TRUE
)
# Bland-Altman plot of all samples mixed
blandAltmanLehPlot(
object = SDDLS,
facet.by = NULL,
color.by = "CellType",
alpha.point = 0.3,
log2 = TRUE
)
 # }
# }